- NengoLoihi
- How to Build a Brain
- Tutorials
- A Single Neuron Model
- Representing a scalar
- Representing a Vector
- Addition
- Arbitrary Linear Transformation
- Nonlinear Transformations
- Structured Representations
- Question Answering
- Question Answering with Control
- Question Answering with Memory
- Learning a communication channel
- Sequencing
- Routed Sequencing
- Routed Sequencing with Cleanup Memory
- Routed Sequencing with Cleanup all Memory
- 2D Decision Integrator
- Requirements
- License
- Tutorials
- NengoCore
- Ablating neurons
- Deep learning
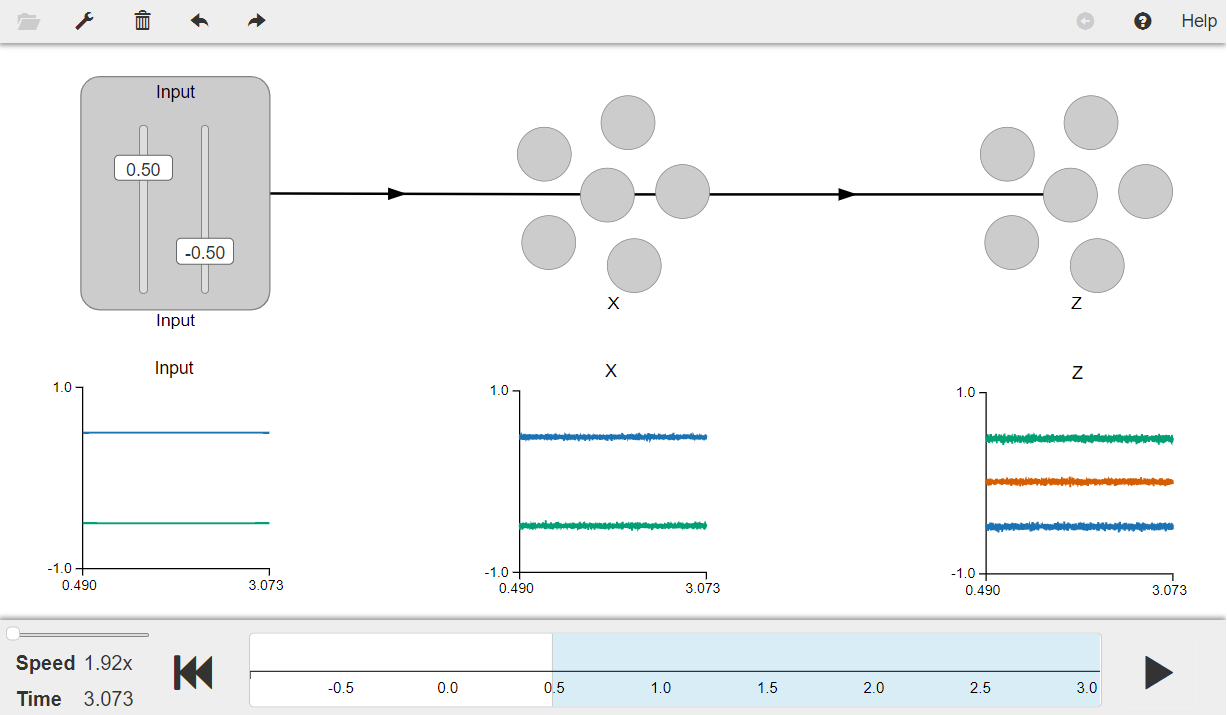
Arbitrary Linear Transformation¶
This model shows that any linear transformation between ensembles can be easily computed by selecting an appropriate value for the “transform”. It provides an example of computing linear transformations using vector representations.
Network diagram:
[Input - 2D] ---> (x - 2D) ---> (z - 3D)
A two-dimensional input signal is first fed into a two-dimensional neuronal ensemble ens_X , which then projects it on to another three-dimensional ensemble ens_Z.
[1]:
# Setup the environment
import nengo
Create the Model¶
This model contains the parameters as described in the book. Setting the transform argument of the connection to be the weight_matrix is analogous to entering the weights in the “2 to 3 Coupling Matrix” window in Nengo 1.4 GUI as described in the book.
[2]:
# Create the network object to which we can add ensembles, connections, etc.
model = nengo.Network(label="Arbitrary Linear Transformation")
with model:
# Two-dimensional input signal with constant value of [0.5, -0.5]
stim = nengo.Node([0.5, -0.5], label="Input")
# 2 and 3-dimensional ensembles each with 200 LIF neurons
ens_X = nengo.Ensemble(200, dimensions=2, label="X")
ens_Z = nengo.Ensemble(200, dimensions=3, label="Z")
# Connect the input to ensemble x
nengo.Connection(stim, ens_X)
# Connect ensemble x to ensemble z using a weight matrix
weight_matrix = [[0.0, 1.0], [1.0, 0.0], [0.5, 0.5]]
nengo.Connection(ens_X, ens_Z, transform=weight_matrix)
Run the Model¶
[ ]:
# Import the nengo_gui visualizer to run and visualize the model.
from nengo_gui.ipython import IPythonViz
IPythonViz(model, "ch3-arbitrary-linear.py.cfg")
Press the play button in the visualizer to run the simulation. You should see the graphs as shown in the figure below.
The graphs show a two-dimesional input linearly projected on to a two-dimensional ensemble of neurons (x), which further linearly projects it on to a three-dimesional neuronal ensemble (z). You can use the sliders to change the input values provided by the input node.
[3]:
from IPython.display import Image
Image(filename="ch3-arbitrary-linear.png")
[3]: